Anand Singh Rathore
I'm Designer
I am a doctoral researcher at the Indraprastha Institute of Information Technology Delhi (IIIT-Delhi), working under the supervision of Prof. Gajendra P.S. Raghava. I hold a master’s degree in Zoology (2020–2022) and have since transitioned into the field of Data Science, driven by the transformative potential of Machine Learning and Artificial Intelligence. My current research focuses on peptide-based therapeutics, particularly exploring their druggability using computational approaches. Computational Biology, being a multidisciplinary field, allows for innovation at the intersection of biology, data science, and artificial intelligence. This fusion offers vast research opportunities to address complex biological challenges and advance therapeutic development.

Zoologist Turned Computational Biologist
Exploring the crossroads of biology and computation to unlock new possibilities in precision medicine and therapeutic design.
- ORCID: 0009-0004-8907-7174
- DOB: 2 July 1998
- Phone: +91 8819919830
- City: New Delhi, India
- Google Scolar: Anand Singh Rathore
- Degree: M.Sc. Zoology
- Email 1: anandr@iiitd.ac.in
- Email 2: anandrathoreindia@gmail.com
I actively work on building and deploying machine learning pipelines for bioinformatics applications such as toxicity prediction, motif analysis, and peptide screening. My toolkit spans Python, R, shell scripting, and technologies like BLAST, IEDB, SwissProt, and ESM-based models. I’m also exploring the frontiers of Quantum Machine Learning and Explainable AI to make bioinformatics models more transparent and insightful. My journey is driven by curiosity, interdisciplinary collaboration, and a desire to make meaningful contributions to biomedical research.
Skills
Comprehensive expertise in bioinformatics programming, data analysis, and computational biology tools.
Resume
Highly motivated research scholar in Computational Biology with strong academic background, practical laboratory training, and diverse experience in machine learning, genomics, and bioinformatics. Committed to contributing to innovative solutions in life sciences through interdisciplinary approaches.
Summary
Education
Ph.D. in Computational Biology (Pursuing)
2022 – Present
Indraprastha Institute of Information Technology (IIIT-Delhi), India
Working under the supervision of Prof. (Dr) GPS Raghava on machine learning applications in peptide-based therapeutics.
M.Sc. in Zoology
2020 – 2022
Hansraj College, University of Delhi, Delhi, India
CGPA: 7.3
Acquired in-depth knowledge in animal physiology, animal endocrinology, molecular biology, developmental biology, and immunology. Gained hands-on experience with biological techniques such as microscopy, gel electrophoresis, and basic bioinformatics tools.
B.Sc. in Life Sciences
2017 – 2020
Sriyut College, APS University, Rewa, Madhyapradesh, India
Percentage: 72.33%
Built a strong foundation in cell biology, genetics, biochemistry, and microbiology. Developed analytical and laboratory skills, and was introduced to research methodologies and scientific writing.
Achievements
CSIR-UGC NET JRF
June 2022
All India Rank: 79 (Percentile: 99.67)
DBT-JRF
2022
Qualified under Category-I
GATE XL
2021
All India Rank: 409 | Score: 598
GATE BT
2022
All India Rank: 906 | Score: 454
Research & Lab Experience
Hands-on Laboratory Training
July – September 2021
Prof. Neeta Sehgal’s Lab, Dept. of Zoology, University of Delhi
- Agarose Gel Electrophoresis, SDS-PAGE, PCR, RNA Isolation
- Western Blotting, Real-Time PCR, DNA & Protein Purification
Antimicrobial Peptide & Database Learning
September – December 2021
Under Asst. Prof. Vikash Sood, Jamia Hamdard
- Peptide data extraction, Venn diagrams, Heatmaps, Clustering
In Silico Biology Virtual Internship
September 2021
Under Dr. Shobhana Manoharan, Biosrishti
- BLAST, FASTA, Phylogenetics, Protein Docking (PatchDock, AutoDock)
- Structure Prediction (Phyre2, SOPMA), Visualization (Chimera, RasMol)
Workshops & Conferences
IEDB Virtual User Workshop
La Jolla Institute for Immunology, USA
Python for Biology Workshop
INSCR 2021
International Conferences
Participated in conferences focusing on Cancer Biology, Nanomedicine, and Genomics
Certifications
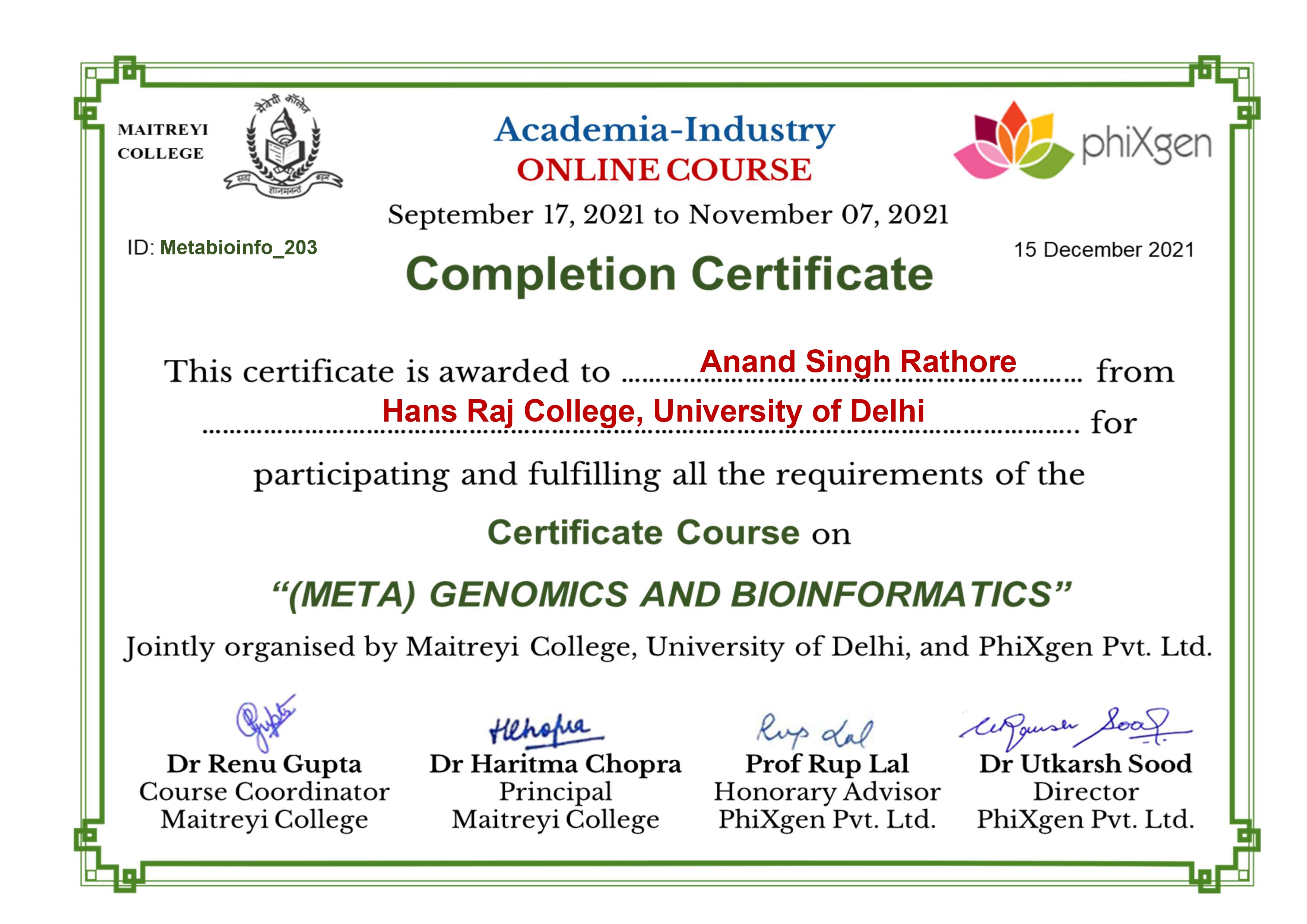
(Meta) Genomics & Bioinformatics
Phixgen Pvt. Ltd.
Immunological Techniques
BCAS, University of Delhi
Secured First Merit Position
Publications
List of published research articles and preprints authored or co-authored by our team.
Published Articles
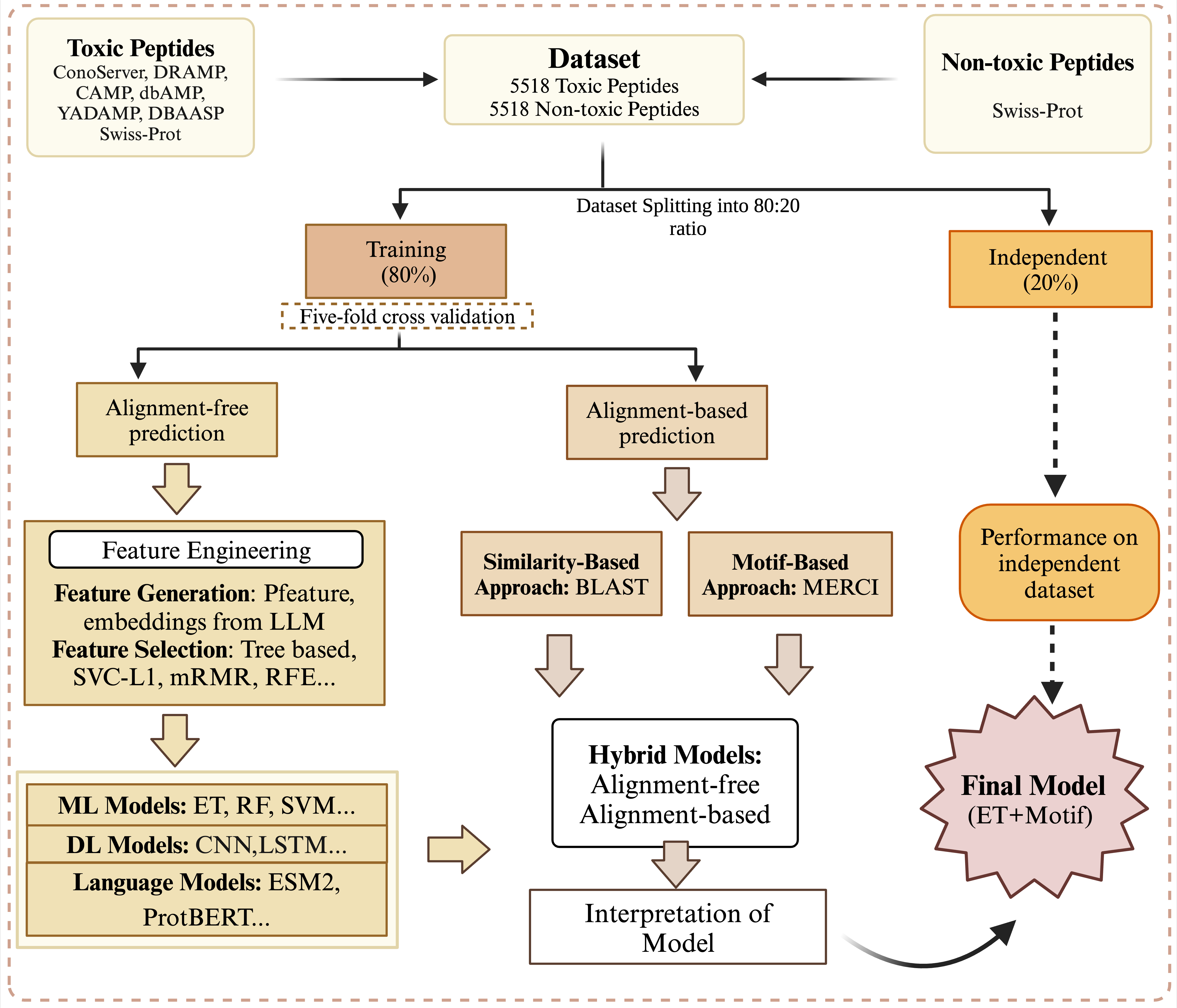
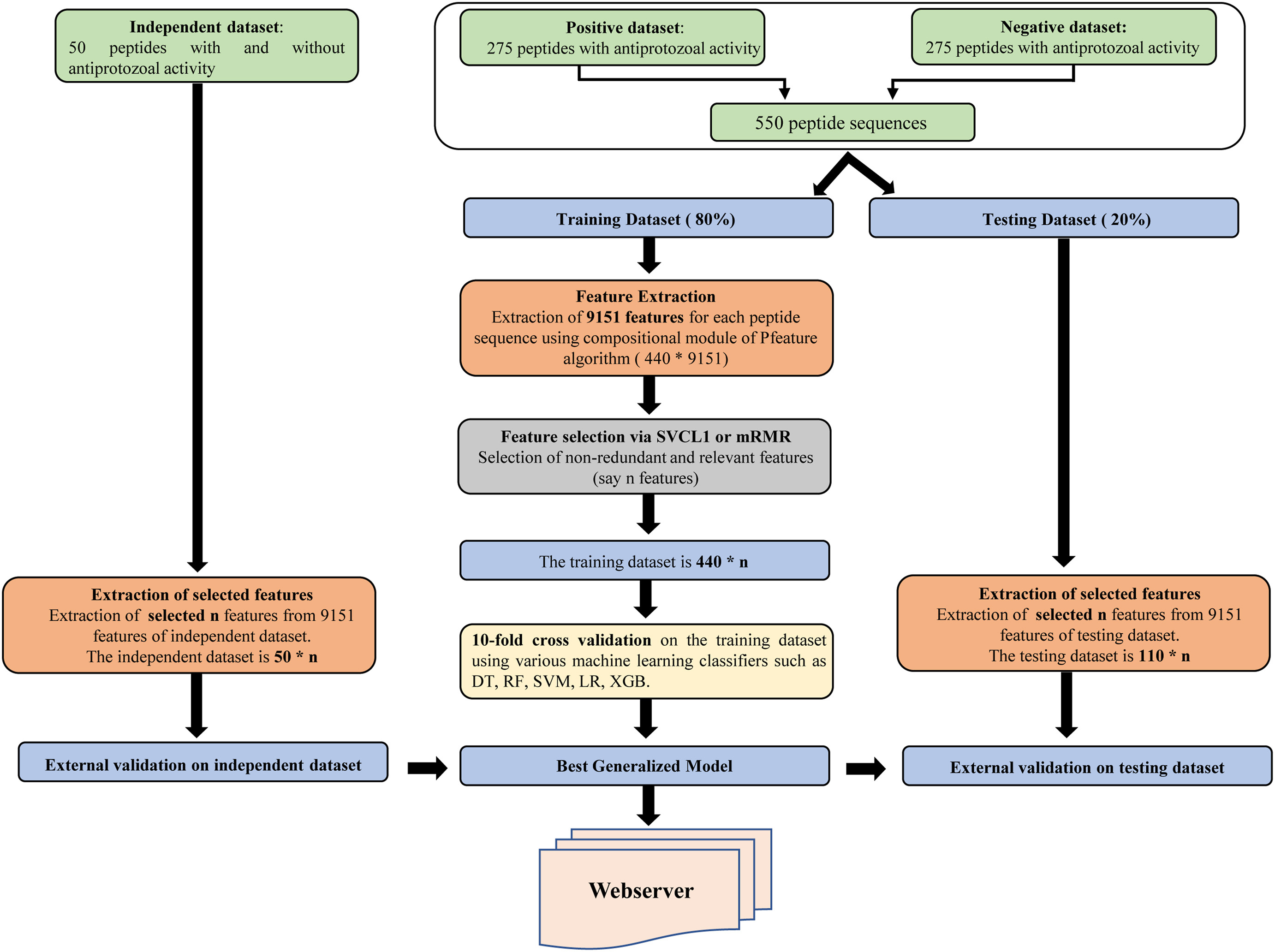
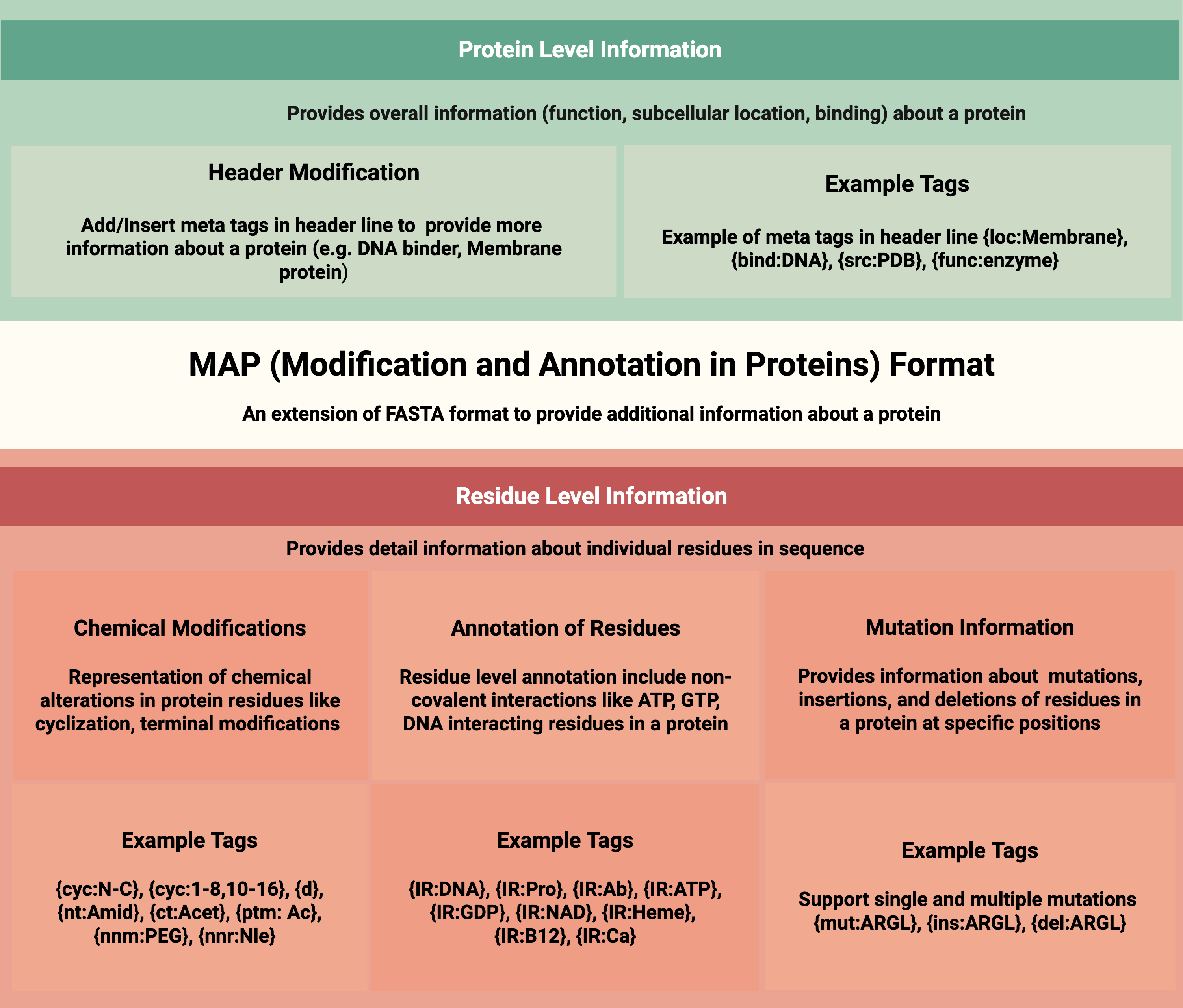
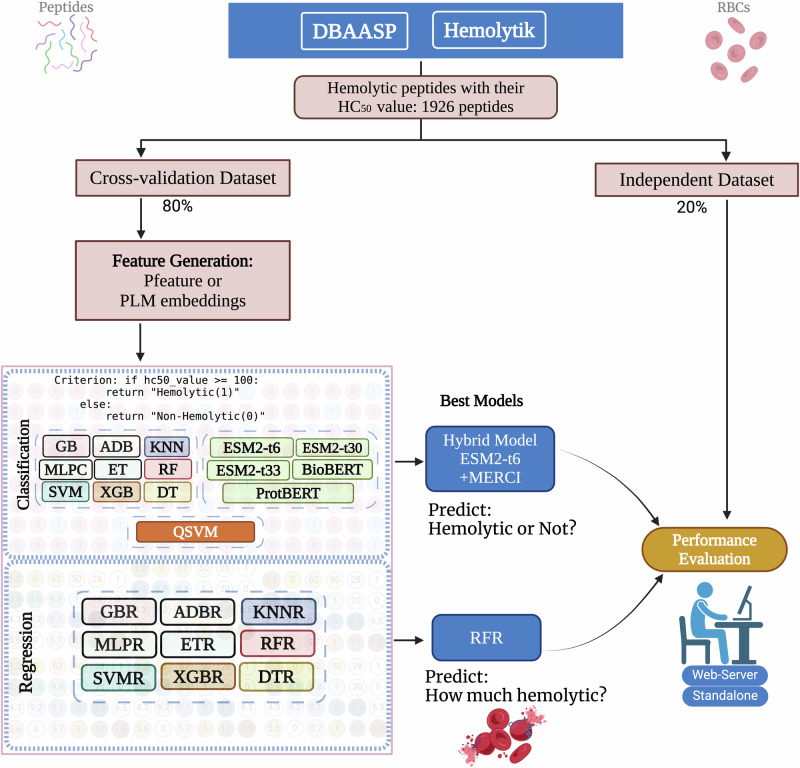
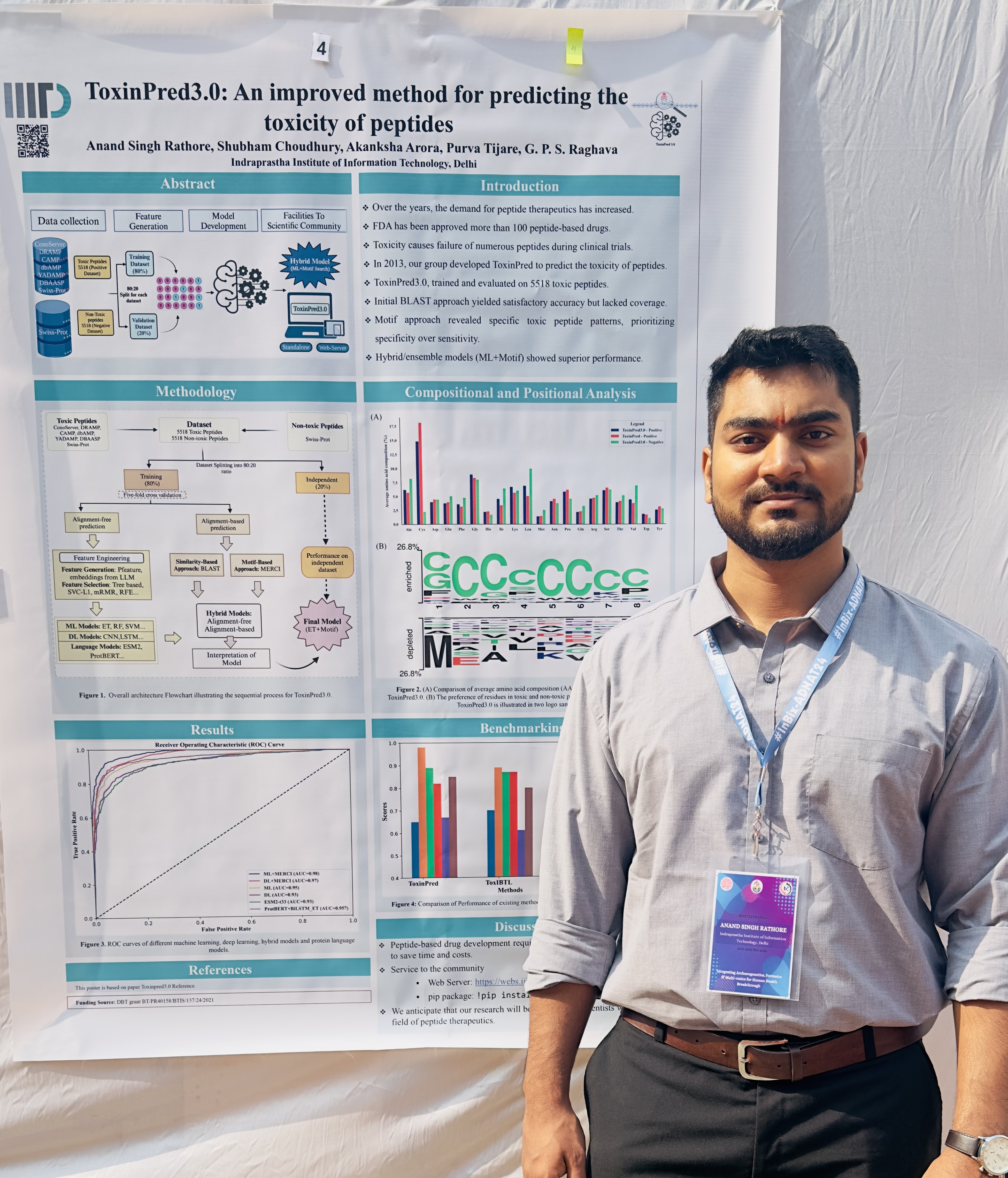
- Rathore AS et al. ToxinPred 3.0: An improved method for predicting...
Computers in Biology and Medicine. 2024; DOI - Rathore AS et al. Prediction of hemolytic peptides...
Communications Biology. 2025; DOI - Rathore AS et al. NTxPred2: A large language model for predicting...
Protein Science. 2025; DOI - Chaudhary S et al. Compilation of resources on subcellular...
Frontiers in RNA Research. 2024; DOI - Periwal N et al. Antiprotozoal peptide prediction using machine...
Heliyon. 2024; DOI
Webservers
This section showcases a collection of web-based resources and tools developed to facilitate peptide-based therapeutic research. These include predictive models, curated databases, and computational methods for analyzing bioactive peptides.
- All
- Tool
- Database
- Method
- Review
- Book Chapter
Research Areas
My research spans a variety of domains in bioinformatics, computational biology, and artificial intelligence to address challenges in health and life sciences.
Machine Learning in Bioinformatics
Development of predictive models for biological data using supervised and unsupervised learning techniques.
Peptide Therapeutics
Design and prediction of therapeutic peptides for antimicrobial, anticancer, and hemolytic activities.
Toxin and Allergen Prediction
Computational tools for the identification of toxic and allergenic proteins and peptides.
Vaccine Informatics
Epitope mapping and computational vaccine design using immunoinformatics approaches.
AI for Precision Medicine
Leveraging artificial intelligence for personalized diagnosis and treatment strategies.
Web Server and Tool Development
Creating user-friendly, freely available web interfaces and software tools for the scientific community's access to predictive models.
Genomics and Sequence Analysis
Exploration of genomic datasets to uncover gene functions, mutations, and their role in disease and evolution.
My Contributions
As a researcher and mentor, I have contributed to multiple facets of academia, individual growth, and broader scientific and societal advancement.
Generation of Knowledge
I developed machine learning and transformer-based models to predict peptide toxicity, neurotoxicity, and hemolytic potential. My work involved curating datasets, designing hybrid frameworks, and deploying accessible tools that enable other researchers to advance peptide-based therapeutics.
Development of Individuals
As a Teaching Assistant, I led tutorials, created content, and mentored students in courses related to biomedical machine learning. I also guided juniors in research methodology and collaborative projects, enhancing their technical and analytical skills.
Contribution to Research Community
I have contributed to interdisciplinary projects, open-source tools, and datasets, while actively engaging in scientific discussions through workshops and conferences. I foster inclusivity and openness in research by sharing resources and supporting diverse collaboration.
Broader Societal Impact
My computational models help in designing safer therapeutic peptides, aligning with public health objectives. I maintain open-access web tools and repositories to ensure these solutions are accessible, encouraging their integration into real-world biomedical pipelines.
Gallery
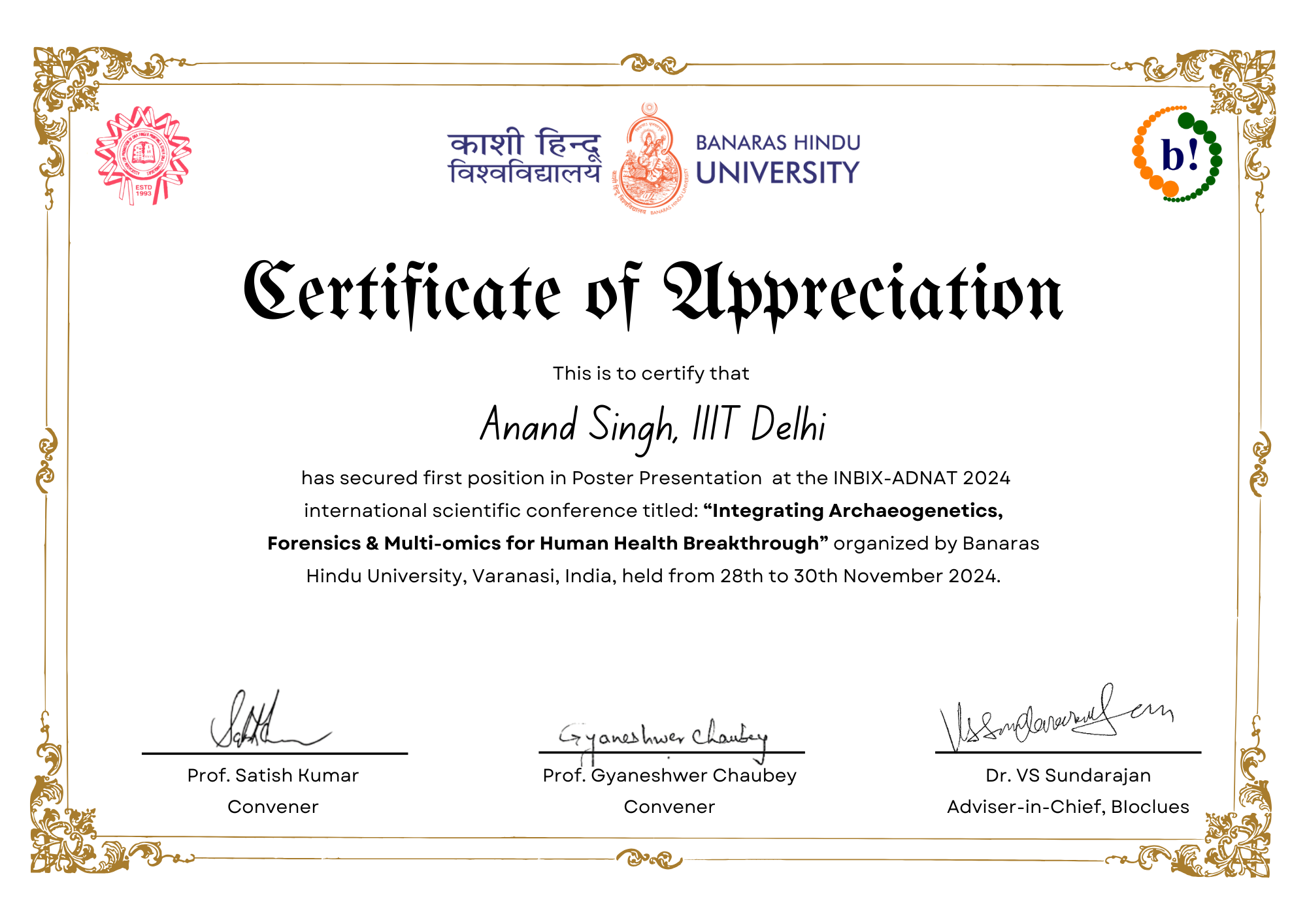
A glimpse into moments, milestones, and memories captured throughout my academic and research journey.
Contact
Feel free to reach out with any questions or collaboration ideas.
Address
A318 R & D Building, IIIT-Delhi
Call
+91 8819919830
anandr@iiitd.ac.in